Effect of deferoxamine on the transcriptome of BSF trypanosomes¶
Michele Tinti & Calvin Tiengwe
set up notebook¶
In [5]:
#reload when modified
%load_ext autoreload
%autoreload 2
#activate r magic
%load_ext rpy2.ipython
%matplotlib inline
In [7]:
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import utilities as UT
import missingno as msno
import random
import matplotlib.pyplot as plt
import matplotlib.patches as patches
import gc
random.seed(1976)
np.random.seed(1976)
Data Anaylsis¶
Experiment SetUp¶
In [8]:
from IPython.display import Image
In [ ]:
In [9]:
#create a dictionary of gene to desc
#from the gff file
def make_desc(_GFF):
gff =pd.read_csv( _GFF, sep='\t', header=None, comment='#')
gff = gff[gff.iloc[:,2]=='gene']
#print( gff[gff[gff.columns[-1]].str.contains('Tb427_020006200')] )
desc = {}
for n in gff.iloc[:,-1]:
n=n.replace('%2C',' ')
item_list = n.split(';')
#print (item_list)
temp_dict = {}
for m in item_list:
#print(m)
temp_dict[m.split('=')[0].strip()]=m.split('=')[1].strip()
#print(temp_dict['ID'])
#print(temp_dict['description'])
desc[temp_dict['ID']]=temp_dict.get('description','none')
return desc
desc_dict = make_desc('genomes/tb927_3/tb927_3.gff')
desc_dict['Tb10.v4.0073']
Out[9]:
Load Read Counts¶
In [13]:
exp = '{life_stage}{replica}'
list_df = [exp.format(
life_stage=life_stage,
replica=replica)
for life_stage in ['C','D','DF','F']
for replica in ['1','2','3']
]
list_df = [n+'/res/'+n+'/counts.txt' for n in list_df]
list_df =[pd.read_csv(n,index_col=[0],comment='#',sep='\t') for n in list_df]
df = list_df[0].copy()
for temp_df in list_df[1:]:
df = df.join(temp_df.iloc[:,-1])
df.head()
#temp_df = pd.read_csv('BSF/tb927_3_ks_counts_final.txt',index_col=[0],comment='#',sep='\t')
Out[13]:
In [14]:
data_col = df.columns[5:]
data_col
Out[14]:
In [15]:
indata = df[data_col]
indata.columns = [n.split('/')[3] for n in indata.columns]
indata.head()
Out[15]:
labels¶
- C - control
- D - cells incubated with iron chelator deferoxamine
- DF - iron-presaturated deferoxamine
- F - FeCl3
In [16]:
print(indata.shape)
indata=indata.dropna()
print(indata.shape)
#indata.loc['KS17gene_1749a']
#indata['desc']=[desc_dict.get(n,'none') for n in indata.index.values]
#indata.to_csv('indata.csv')
#indata.head()
#indata.loc['mainVSG-427-2']
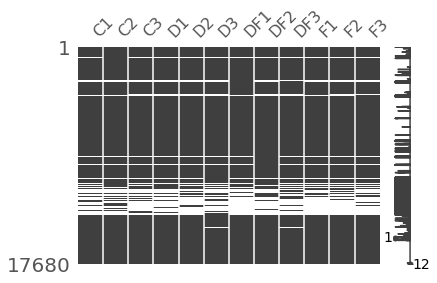
Missing Data Viz¶
In [ ]:
In [17]:
msno.matrix(indata.replace(0,np.nan).dropna(how='all'),figsize=(6, 4))
Out[17]:
In [18]:
msno.bar(indata.replace(0,np.nan).dropna(how='all'),figsize=(6, 4))
Out[18]:
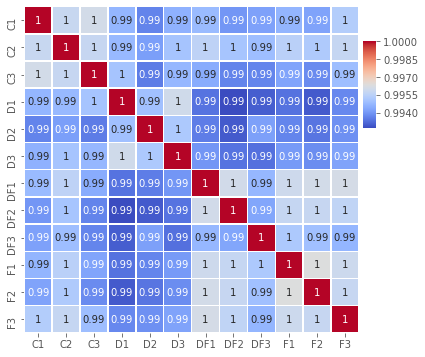
QC - Corr analysis¶
In [19]:
!mkdir -p Figures
In [20]:
fig,ax=plt.subplots(figsize=(6,6))
cbar_ax = fig.add_axes([.91, .6, .03, .2])
sns.heatmap(np.log2(indata).corr(),
#vmin=-1,
cmap='coolwarm',
annot=True,linewidths=.5,ax=ax, cbar_ax = cbar_ax, cbar=True)
print(ax.get_ylim())
ax.set_ylim(12,0)
plt.savefig('Figures/Figure_2.png')
plt.show()
In [ ]:
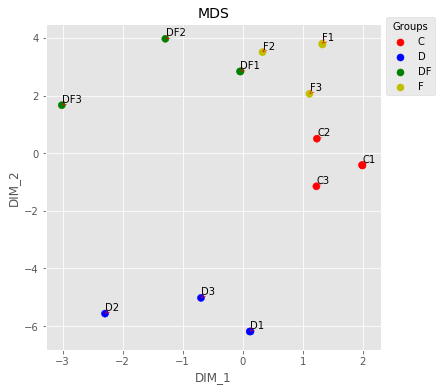
QC - MSD¶
In [21]:
plt.style.use('ggplot')
palette = ['r']*3+['b']*3+['g']*3+['y']*3
fig,ax = plt.subplots(figsize=(6,6), ncols=1, nrows=1)
UT.make_mds(np.log2(indata),palette,ax,color_dictionary={'r':'C','b':'D',
'g':'DF','y':'F',})
plt.savefig('Figures/Figure_3.png')
plt.show()
In [20]:
def get_gene_ids(n):
res = {}
temp = n.split(';')
temp =[n.strip() for n in temp if len(n)>2]
for f in temp:
key = f.split(' ')[0]
value = f.split(' ')[1]
key=key.replace('\"','').replace('\'','').strip()
value=value.replace('\"','').replace('\'','').strip()
res[key]=value
return res['gene_id']
edgeR analysis¶
In [22]:
%%R -i indata
options(warn=-1)
library("limma")
library("edgeR")
head(indata)
In [23]:
%%R
group <- factor(c(
'C','C','C',
'D','D','D',
'DF','DF','DF',
'F','F','F'
))
design_with_all <- model.matrix( ~0+group )
y <- DGEList(counts=indata,group=group)
keep <- filterByExpr(y)
y <- y[keep,,keep.lib.sizes=FALSE]
counts = y$counts
genes = row.names(y)
y <- calcNormFactors(y)
y <- estimateDisp(y, design_with_all)
fit_all <- glmQLFit( y, design_with_all )
Differential Expression Analysis¶
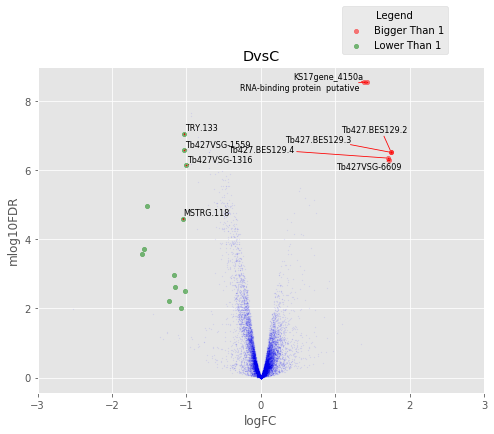
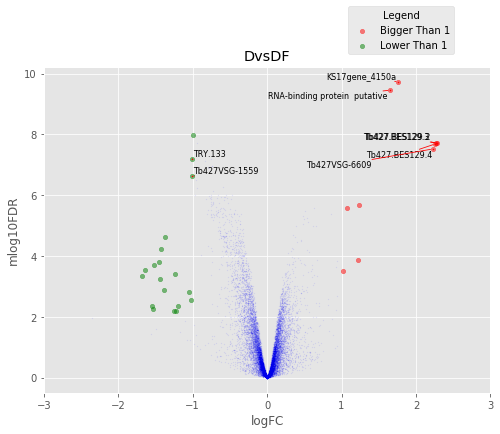
D vs C¶
In [28]:
%%R
contrastDC <- glmQLFTest( fit_all, contrast=makeContrasts( groupD-groupC, levels=design_with_all ) )
tableDC <- topTags(contrastDC, n=Inf, sort.by = "none", adjust.method="BH")$table
head(tableDC)
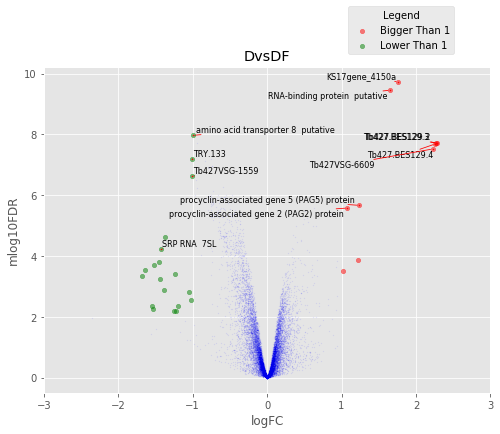
D vs DF¶
In [29]:
%%R
contrastDDF <- glmQLFTest( fit_all, contrast=makeContrasts( groupD-groupDF, levels=design_with_all ) )
tableDDF <- topTags(contrastDDF, n=Inf, sort.by = "none", adjust.method="BH")$table
head(tableDDF)
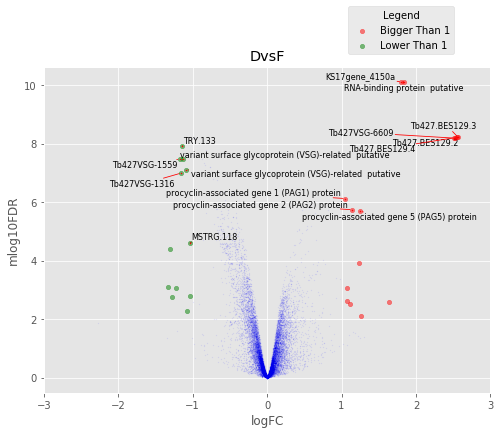
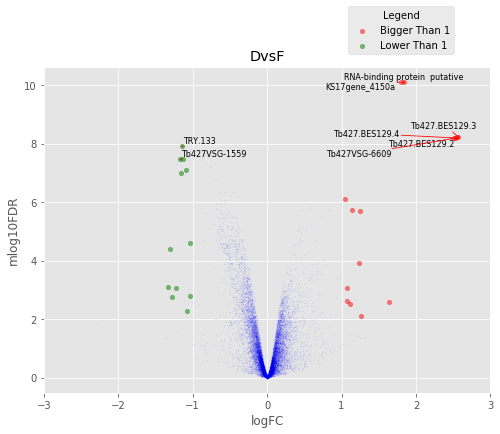
D vs F¶
In [30]:
%%R
contrastDF <- glmQLFTest( fit_all, contrast=makeContrasts( groupD-groupF, levels=design_with_all ) )
tableDF <- topTags(contrastDF, n=Inf, sort.by = "none", adjust.method="BH")$table
head(tableDF)
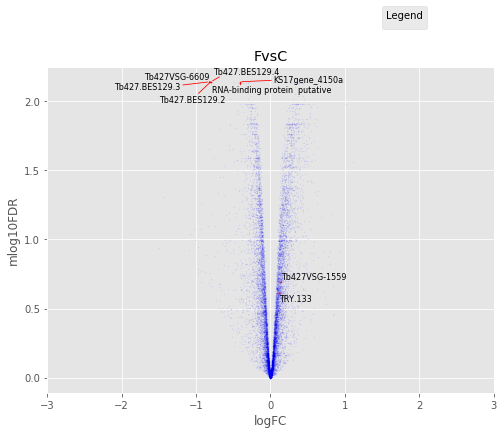
F vs C¶
In [31]:
%%R
contrastFC <- glmQLFTest( fit_all, contrast=makeContrasts( groupF-groupC, levels=design_with_all ) )
tableFC <- topTags(contrastFC, n=Inf, sort.by = "none", adjust.method="BH")$table
head(tableFC)
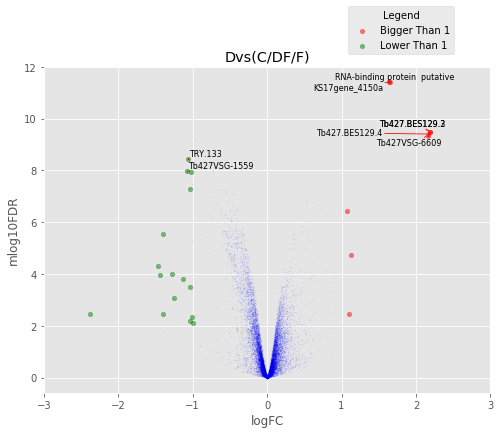
D vs (DF+C+F)/3¶
In [32]:
%%R
contrast_mine <- glmQLFTest( fit_all, contrast=makeContrasts( groupD-(groupDF+groupC+groupF)/3, levels=design_with_all ) )
table_mine <- topTags(contrast_mine, n=Inf, sort.by = "none", adjust.method="BH")$table
head(table_mine)
In [33]:
%R -o tableDC,tableDDF,tableDF,tableFC,table_mine
def mod_table(intable):
intable['mlog10FDR']=-np.log10(intable['FDR'])
intable['mlog10pvalue']=-np.log10(intable['PValue'])
intable['desc']=[desc_dict.get(n,n) for n in intable.index.values]
return intable
tableDC = mod_table(tableDC)
tableDDF = mod_table(tableDDF)
tableDF = mod_table(tableDF)
tableFC = mod_table(tableFC)
table_mine = mod_table(table_mine)
Exploratory Pltots¶
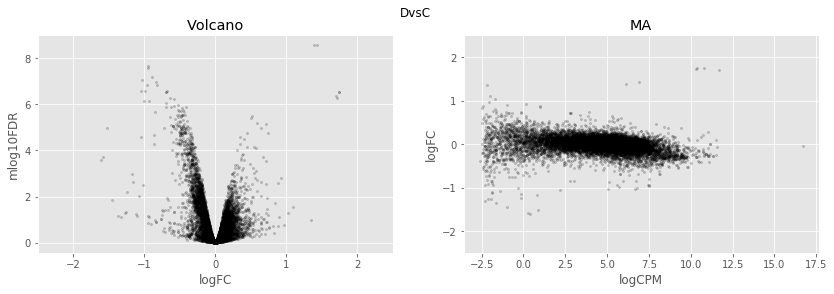
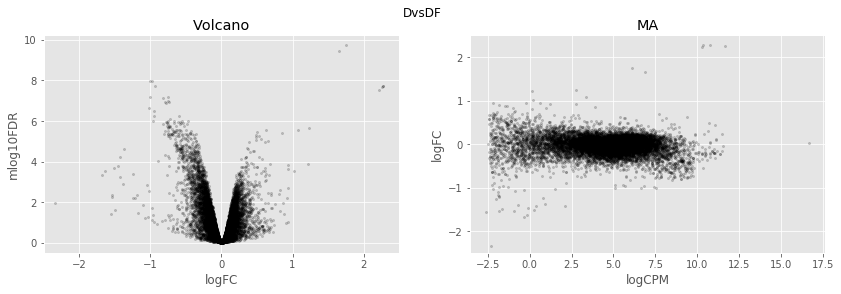
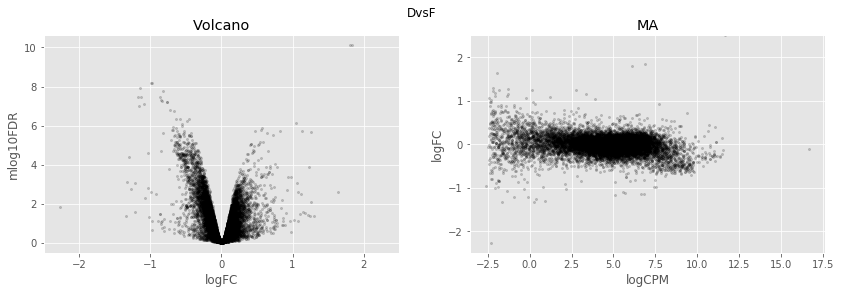
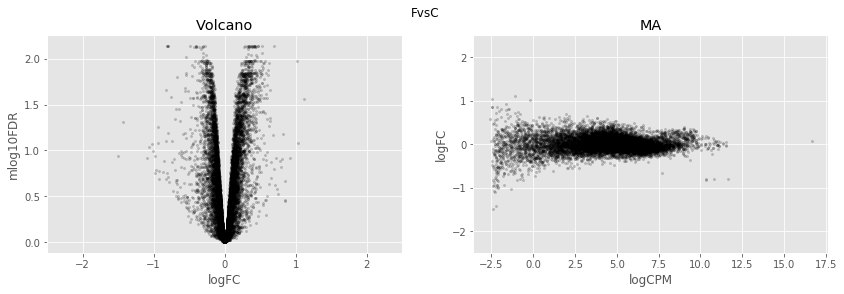
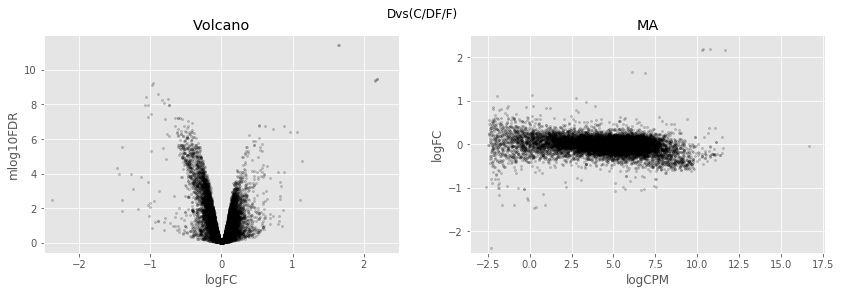
In [34]:
for table,name in zip([tableDC,tableDDF,tableDF,tableFC,table_mine],
['DvsC','DvsDF','DvsF','FvsC','Dvs(C/DF/F)']):
fig,axes=plt.subplots(figsize=(14,4), ncols=2, nrows=1)
ax=axes[0]
table.plot(x='logFC',y='mlog10FDR',
kind='scatter',s=5,alpha=0.2,ax=ax,c='black')
ax.set_xlim(-2.5,2.5)
ax.set_title('Volcano')
ax=axes[1]
table.plot(x='logCPM',y='logFC',
kind='scatter',s=5,alpha=0.2,ax=ax,c='black')
ax.set_ylim(-2.5,2.5)
ax.set_title('MA')
plt.suptitle(name)
plt.show()
In [35]:
def annotated_volcano(table,name,selection=False):
fig,axes=plt.subplots(figsize=(8,6), ncols=1, nrows=1)
ax=axes
if not selection:
selection = table[((table['logFC']>1)|(table['logFC']<-1))
&(table['mlog10FDR']>2)&(table['logCPM']>2)]
else:
selection = table.loc[selection]
annot_index = selection.index.values
annot_names = selection['desc']
UT.make_vulcano(
table,
ax,
x='logFC',
y='mlog10FDR',
fc_col = 'logFC',
pval_col = 'FDR',
pval_limit=0.01,
fc_limit=1,
annot_index=annot_index ,
annot_names=annot_names)
ax.legend(loc='upper center', bbox_to_anchor=(0.8, 1.2), title='Legend')
ax.set_xlim(-3,3)
plt.title(name)
Volcano Plots¶
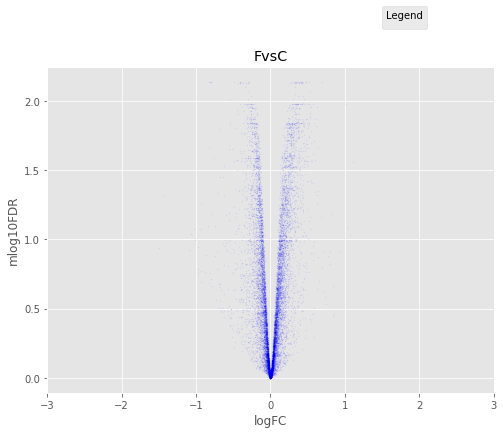
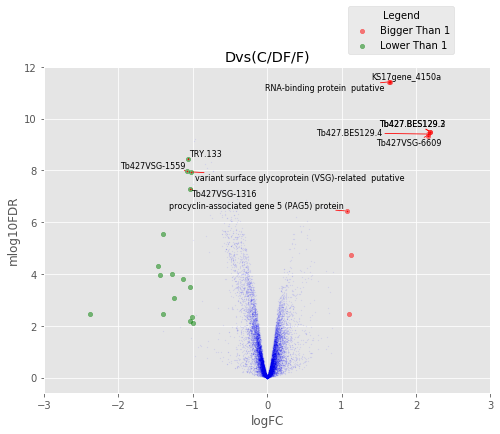
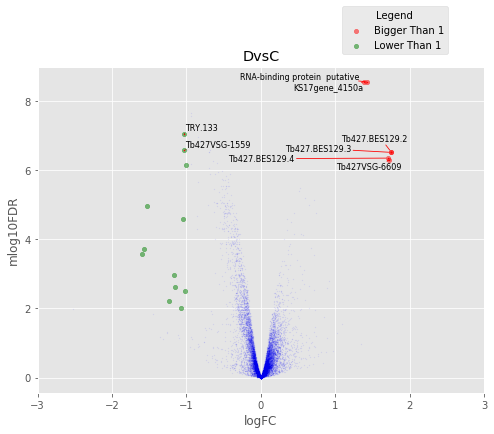
In [37]:
for table,name in zip([tableDC,tableDDF,tableDF,tableFC,table_mine],
['DvsC','DvsDF','DvsF','FvsC','Dvs(C/DF/F)']):
annotated_volcano(table,name)
In [41]:
table_dict = {}
for table,name in zip([tableDC,tableDDF,tableDF],['tableDC','tableDDF','tableDF']):
selection = table[((table['logFC']>1)|(table['logFC']<-1))
&(table['mlog10FDR']>2)&(table['logCPM']>1)]
table_dict[name]= list(selection.index.values)
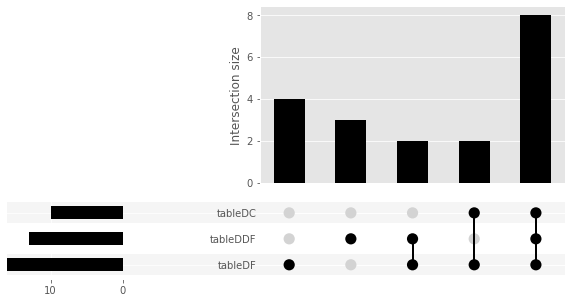
Upset Plots¶
To identify differentially regulated genes in common between the conditions
In [42]:
from upsetplot import plot,UpSet
from upsetplot import from_contents
import matplotlib
data_upset = from_contents(table_dict)
plot(data_upset)
fig = plt.gcf()
fig.set_size_inches(10, 5)
plt.show()
In [46]:
#list of common genes
common = set(table_dict['tableDC']) & set(table_dict['tableDDF']) & set(table_dict['tableDF'])
common
Out[46]:
Visulize the common proteins¶
In [45]:
for table,name in zip([tableDC,tableDDF,tableDF,tableFC,table_mine],
['DvsC','DvsDF','DvsF','FvsC','Dvs(C/DF/F)']):
annotated_volcano(table,name,selection=list(common))
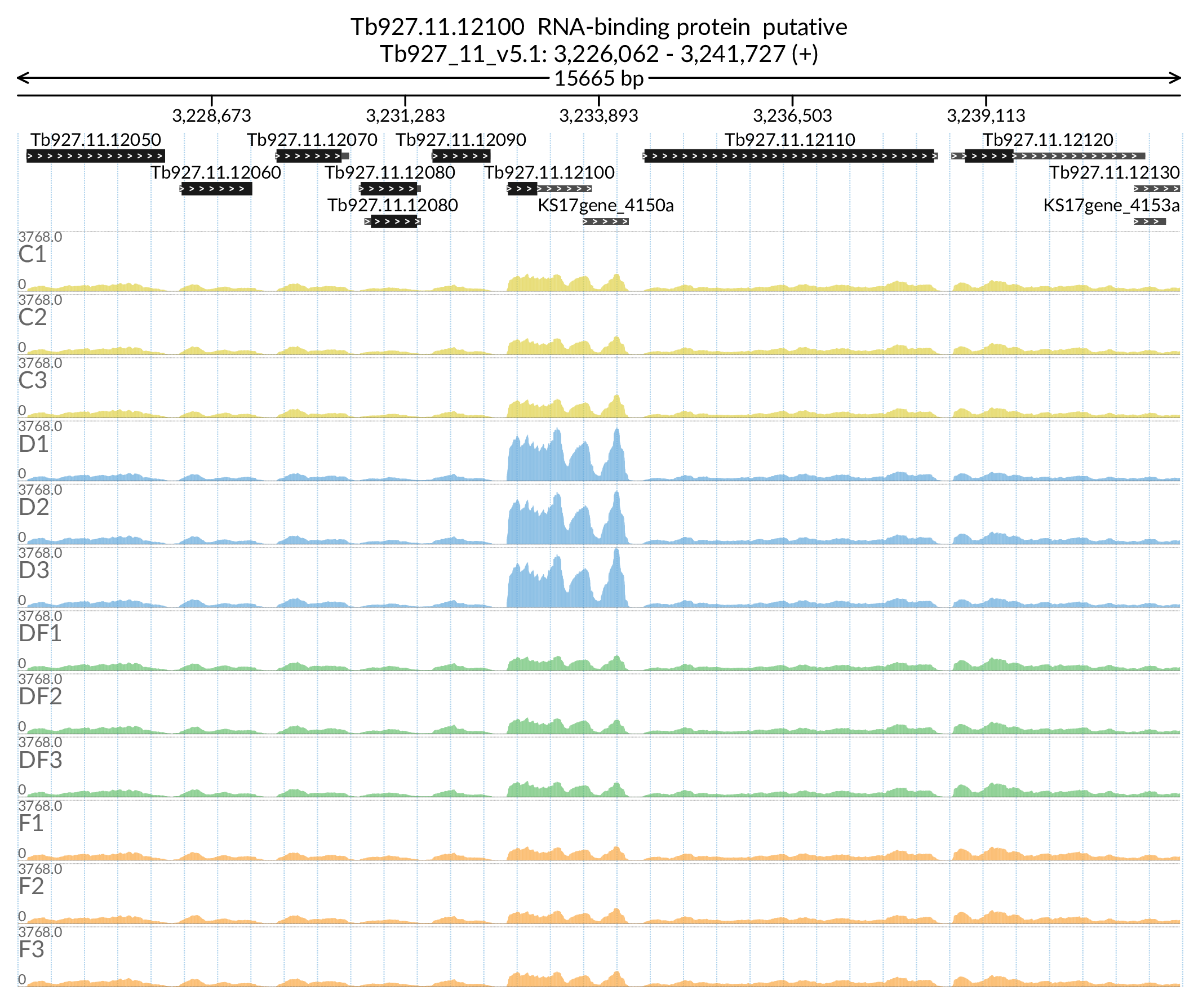
In [47]:
Image('Figures/Tb927.11.12100_paperFig.png')
Out[47]:
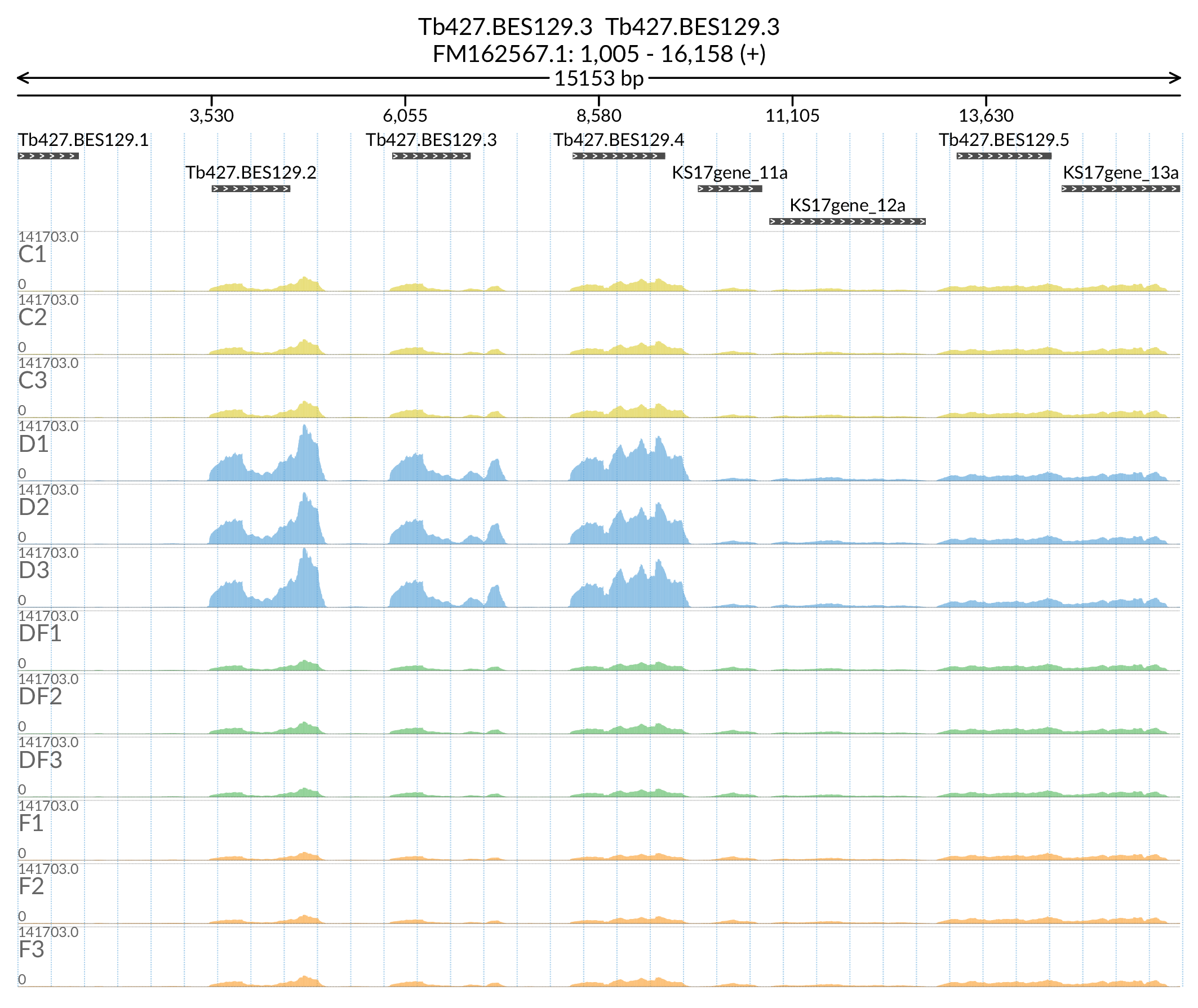
In [50]:
Image('Figures/Tb427.BES129.3_paperFig.png')
Out[50]:
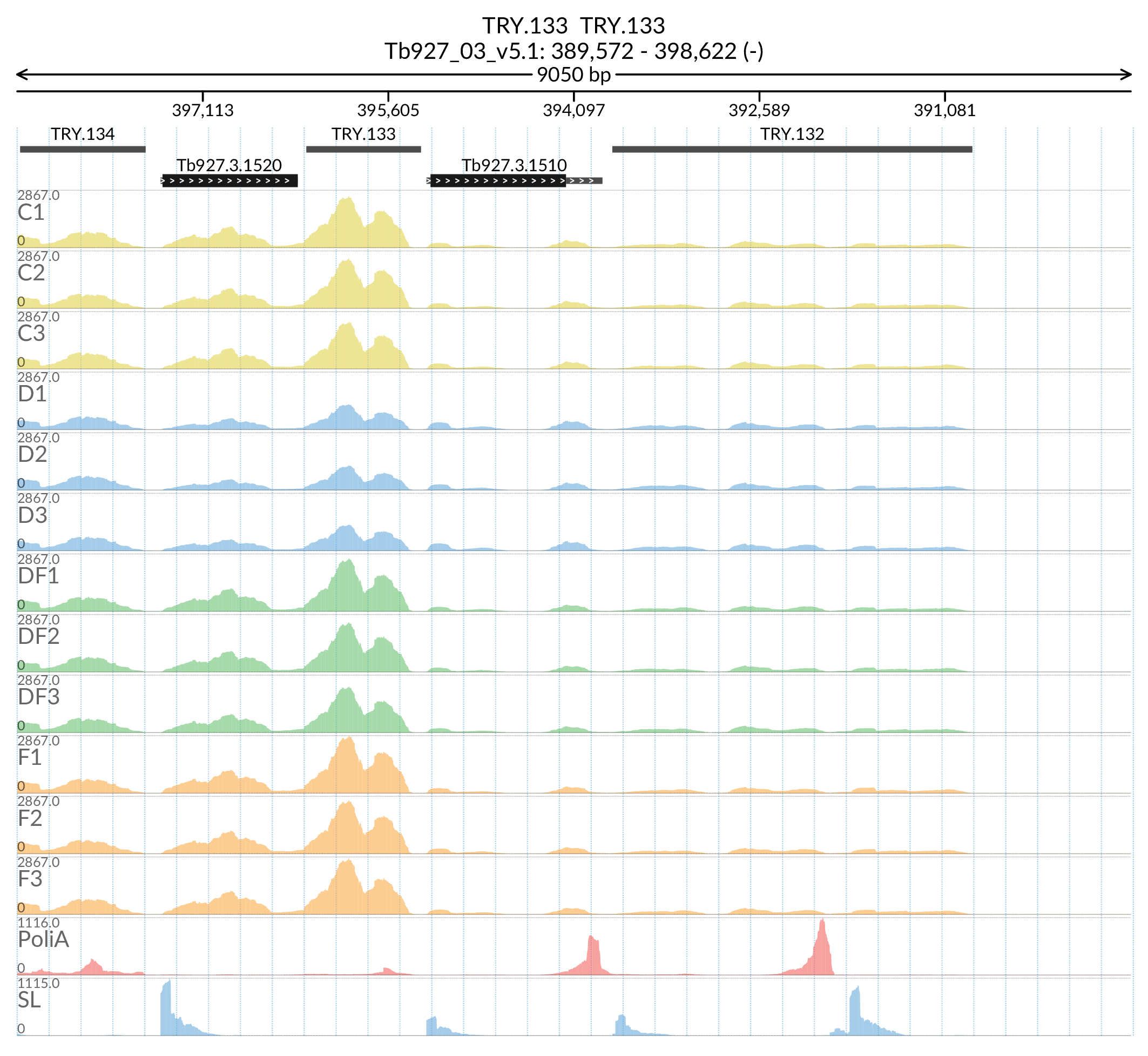
In [53]:
Image('Figures/TRY.133_paperFigSL.png')
Out[53]:
In [ ]:
!jupyter nbconvert --to html_toc analysis_def.ipynb